
A | B | C | D | E | F | G | H | CH | I | J | K | L | M | N | O | P | Q | R | S | T | U | V | W | X | Y | Z | 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9
| Part of a series on |
| Genetics |
|---|
 |
Deoxyribonucleic acid (/diːˈɒksɪˌraɪboʊnjuːˌkliːɪk, -ˌkleɪ-/ ;[1] DNA) is a polymer composed of two polynucleotide chains that coil around each other to form a double helix. The polymer carries genetic instructions for the development, functioning, growth and reproduction of all known organisms and many viruses. DNA and ribonucleic acid (RNA) are nucleic acids. Alongside proteins, lipids and complex carbohydrates (polysaccharides), nucleic acids are one of the four major types of macromolecules that are essential for all known forms of life.
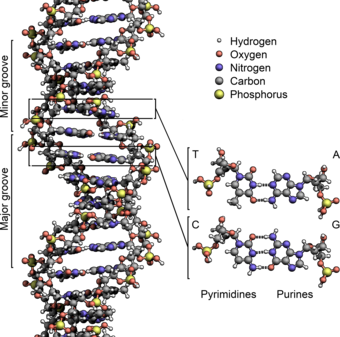
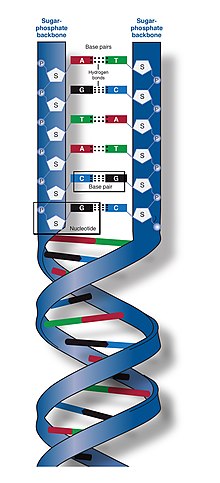
The two DNA strands are known as polynucleotides as they are composed of simpler monomeric units called nucleotides.[2][3] Each nucleotide is composed of one of four nitrogen-containing nucleobases (cytosine , guanine , adenine or thymine ), a sugar called deoxyribose, and a phosphate group. The nucleotides are joined to one another in a chain by covalent bonds (known as the phosphodiester linkage) between the sugar of one nucleotide and the phosphate of the next, resulting in an alternating sugar-phosphate backbone. The nitrogenous bases of the two separate polynucleotide strands are bound together, according to base pairing rules (A with T and C with G), with hydrogen bonds to make double-stranded DNA. The complementary nitrogenous bases are divided into two groups, the single-ringed pyrimidines and the double-ringed purines. In DNA, the pyrimidines are thymine and cytosine; the purines are adenine and guanine.
Both strands of double-stranded DNA store the same biological information. This information is replicated when the two strands separate. A large part of DNA (more than 98% for humans) is non-coding, meaning that these sections do not serve as patterns for protein sequences. The two strands of DNA run in opposite directions to each other and are thus antiparallel. Attached to each sugar is one of four types of nucleobases (or bases). It is the sequence of these four nucleobases along the backbone that encodes genetic information. RNA strands are created using DNA strands as a template in a process called transcription, where DNA bases are exchanged for their corresponding bases except in the case of thymine (T), for which RNA substitutes uracil (U).[4] Under the genetic code, these RNA strands specify the sequence of amino acids within proteins in a process called translation.
Within eukaryotic cells, DNA is organized into long structures called chromosomes. Before typical cell division, these chromosomes are duplicated in the process of DNA replication, providing a complete set of chromosomes for each daughter cell. Eukaryotic organisms (animals, plants, fungi and protists) store most of their DNA inside the cell nucleus as nuclear DNA, and some in the mitochondria as mitochondrial DNA or in chloroplasts as chloroplast DNA.[5] In contrast, prokaryotes (bacteria and archaea) store their DNA only in the cytoplasm, in circular chromosomes. Within eukaryotic chromosomes, chromatin proteins, such as histones, compact and organize DNA. These compacting structures guide the interactions between DNA and other proteins, helping control which parts of the DNA are transcribed.
Properties
DNA is a long polymer made from repeating units called nucleotides.[6][7] The structure of DNA is dynamic along its length, being capable of coiling into tight loops and other shapes.[8] In all species it is composed of two helical chains, bound to each other by hydrogen bonds. Both chains are coiled around the same axis, and have the same pitch of 34 ångströms (3.4 nm). The pair of chains have a radius of 10 Å (1.0 nm).[9] According to another study, when measured in a different solution, the DNA chain measured 22–26 Å (2.2–2.6 nm) wide, and one nucleotide unit measured 3.3 Å (0.33 nm) long.[10] The buoyant density of most DNA is 1.7g/cm3.[11]
DNA does not usually exist as a single strand, but instead as a pair of strands that are held tightly together.[9][12] These two long strands coil around each other, in the shape of a double helix. The nucleotide contains both a segment of the backbone of the molecule (which holds the chain together) and a nucleobase (which interacts with the other DNA strand in the helix). A nucleobase linked to a sugar is called a nucleoside, and a base linked to a sugar and to one or more phosphate groups is called a nucleotide. A biopolymer comprising multiple linked nucleotides (as in DNA) is called a polynucleotide.[13]
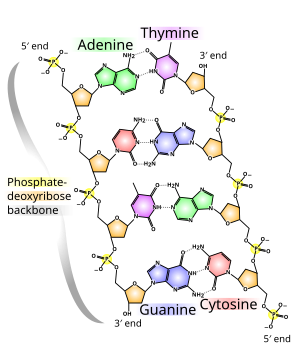
The backbone of the DNA strand is made from alternating phosphate and sugar groups.[14] The sugar in DNA is 2-deoxyribose, which is a pentose (five-carbon) sugar. The sugars are joined by phosphate groups that form phosphodiester bonds between the third and fifth carbon atoms of adjacent sugar rings. These are known as the 3′-end (three prime end), and 5′-end (five prime end) carbons, the prime symbol being used to distinguish these carbon atoms from those of the base to which the deoxyribose forms a glycosidic bond.[12]
Therefore, any DNA strand normally has one end at which there is a phosphate group attached to the 5′ carbon of a ribose (the 5′ phosphoryl) and another end at which there is a free hydroxyl group attached to the 3′ carbon of a ribose (the 3′ hydroxyl). The orientation of the 3′ and 5′ carbons along the sugar-phosphate backbone confers directionality (sometimes called polarity) to each DNA strand. In a nucleic acid double helix, the direction of the nucleotides in one strand is opposite to their direction in the other strand: the strands are antiparallel. The asymmetric ends of DNA strands are said to have a directionality of five prime end (5′ ), and three prime end (3′), with the 5′ end having a terminal phosphate group and the 3′ end a terminal hydroxyl group. One major difference between DNA and RNA is the sugar, with the 2-deoxyribose in DNA being replaced by the related pentose sugar ribose in RNA.[12]
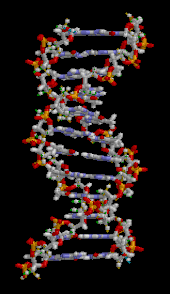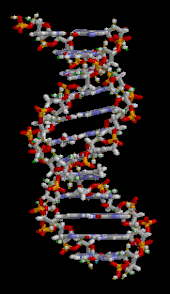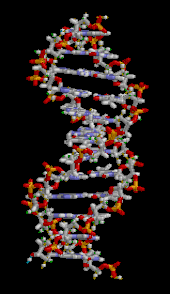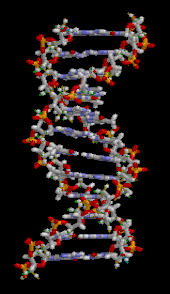
The DNA double helix is stabilized primarily by two forces: hydrogen bonds between nucleotides and base-stacking interactions among aromatic nucleobases.[16] The four bases found in DNA are adenine (A), cytosine (C), guanine (G) and thymine (T). These four bases are attached to the sugar-phosphate to form the complete nucleotide, as shown for adenosine monophosphate. Adenine pairs with thymine and guanine pairs with cytosine, forming A-T and G-C base pairs.[17][18]
Nucleobase classification
The nucleobases are classified into two types: the purines, A and G, which are fused five- and six-membered heterocyclic compounds, and the pyrimidines, the six-membered rings C and T.[12] A fifth pyrimidine nucleobase, uracil (U), usually takes the place of thymine in RNA and differs from thymine by lacking a methyl group on its ring. In addition to RNA and DNA, many artificial nucleic acid analogues have been created to study the properties of nucleic acids, or for use in biotechnology.[19]
Non-canonical bases
Modified bases occur in DNA. The first of these recognized was 5-methylcytosine, which was found in the genome of Mycobacterium tuberculosis in 1925.[20] The reason for the presence of these noncanonical bases in bacterial viruses (bacteriophages) is to avoid the restriction enzymes present in bacteria. This enzyme system acts at least in part as a molecular immune system protecting bacteria from infection by viruses.[21] Modifications of the bases cytosine and adenine, the more common and modified DNA bases, play vital roles in the epigenetic control of gene expression in plants and animals.[22]
A number of noncanonical bases are known to occur in DNA.[23] Most of these are modifications of the canonical bases plus uracil.
- Modified Adenine
- N6-carbamoyl-methyladenine
- N6-methyadenine
- Modified Guanine
- 7-Deazaguanine
- 7-Methylguanine
- Modified Cytosine
- N4-Methylcytosine
- 5-Carboxylcytosine
- 5-Formylcytosine
- 5-Glycosylhydroxymethylcytosine
- 5-Hydroxycytosine
- 5-Methylcytosine
- Modified Thymidine
- α-Glutamythymidine
- α-Putrescinylthymine
- Uracil and modifications
- Base J
- Uracil
- 5-Dihydroxypentauracil
- 5-Hydroxymethyldeoxyuracil
- Others
- Deoxyarchaeosine
- 2,6-Diaminopurine (2-Aminoadenine)
Grooves
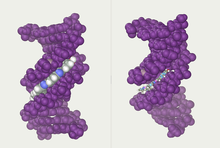
Twin helical strands form the DNA backbone. Another double helix may be found tracing the spaces, or grooves, between the strands. These voids are adjacent to the base pairs and may provide a binding site. As the strands are not symmetrically located with respect to each other, the grooves are unequally sized. The major groove is 22 ångströms (2.2 nm) wide, while the minor groove is 12 Å (1.2 nm) in width.[24] Due to the larger width of the major groove, the edges of the bases are more accessible in the major groove than in the minor groove. As a result, proteins such as transcription factors that can bind to specific sequences in double-stranded DNA usually make contact with the sides of the bases exposed in the major groove.[25] This situation varies in unusual conformations of DNA within the cell (see below), but the major and minor grooves are always named to reflect the differences in width that would be seen if the DNA was twisted back into the ordinary B form.
Base pairing

|
|
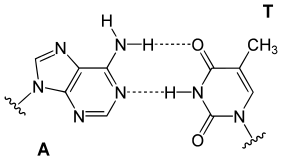
In a DNA double helix, each type of nucleobase on one strand bonds with just one type of nucleobase on the other strand. This is called complementary base pairing. Purines form hydrogen bonds to pyrimidines, with adenine bonding only to thymine in two hydrogen bonds, and cytosine bonding only to guanine in three hydrogen bonds. This arrangement of two nucleotides binding together across the double helix (from six-carbon ring to six-carbon ring) is called a Watson-Crick base pair. DNA with high GC-content is more stable than DNA with low GC-content. A Hoogsteen base pair (hydrogen bonding the 6-carbon ring to the 5-carbon ring) is a rare variation of base-pairing.[26] As hydrogen bonds are not covalent, they can be broken and rejoined relatively easily. The two strands of DNA in a double helix can thus be pulled apart like a zipper, either by a mechanical force or high temperature.[27] As a result of this base pair complementarity, all the information in the double-stranded sequence of a DNA helix is duplicated on each strand, which is vital in DNA replication. This reversible and specific interaction between complementary base pairs is critical for all the functions of DNA in organisms.[7]
ssDNA vs. dsDNA
Most DNA molecules are actually two polymer strands, bound together in a helical fashion by noncovalent bonds; this double-stranded (dsDNA) structure is maintained largely by the intrastrand base stacking interactions, which are strongest for G,C stacks. The two strands can come apart—a process known as melting—to form two single-stranded DNA (ssDNA) molecules. Melting occurs at high temperatures, low salt and high pH (low pH also melts DNA, but since DNA is unstable due to acid depurination, low pH is rarely used).
The stability of the dsDNA form depends not only on the GC-content (% G,C basepairs) but also on sequence (since stacking is sequence specific) and also length (longer molecules are more stable). The stability can be measured in various ways; a common way is the melting temperature (also called Tm value), which is the temperature at which 50% of the double-strand molecules are converted to single-strand molecules; melting temperature is dependent on ionic strength and the concentration of DNA. As a result, it is both the percentage of GC base pairs and the overall length of a DNA double helix that determines the strength of the association between the two strands of DNA. Long DNA helices with a high GC-content have more strongly interacting strands, while short helices with high AT content have more weakly interacting strands.[28] In biology, parts of the DNA double helix that need to separate easily, such as the TATAAT Pribnow box in some promoters, tend to have a high AT content, making the strands easier to pull apart.[29]
In the laboratory, the strength of this interaction can be measured by finding the melting temperature Tm necessary to break half of the hydrogen bonds. When all the base pairs in a DNA double helix melt, the strands separate and exist in solution as two entirely independent molecules. These single-stranded DNA molecules have no single common shape, but some conformations are more stable than others.[30]
Amount

In humans, the total female diploid nuclear genome per cell extends for 6.37 Gigabase pairs (Gbp), is 208.23 cm long and weighs 6.51 picograms (pg).[31] Male values are 6.27 Gbp, 205.00 cm, 6.41 pg.[31] Each DNA polymer can contain hundreds of millions of nucleotides, such as in chromosome 1. Chromosome 1 is the largest human chromosome with approximately 220 million base pairs, and would be 85 mm long if straightened.[32]
In eukaryotes, in addition to nuclear DNA, there is also mitochondrial DNA (mtDNA) which encodes certain proteins used by the mitochondria. The mtDNA is usually relatively small in comparison to the nuclear DNA. For example, the human mitochondrial DNA forms closed circular molecules, each of which contains 16,569[33][34] DNA base pairs,[35] with each such molecule normally containing a full set of the mitochondrial genes. Each human mitochondrion contains, on average, approximately 5 such mtDNA molecules.[35] Each human cell contains approximately 100 mitochondria, giving a total number of mtDNA molecules per human cell of approximately 500.[35] However, the amount of mitochondria per cell also varies by cell type, and an egg cell can contain 100,000 mitochondria, corresponding to up to 1,500,000 copies of the mitochondrial genome (constituting up to 90% of the DNA of the cell).[36]
Sense and antisense
A DNA sequence is called a "sense" sequence if it is the same as that of a messenger RNA copy that is translated into protein.[37] The sequence on the opposite strand is called the "antisense" sequence. Both sense and antisense sequences can exist on different parts of the same strand of DNA (i.e. both strands can contain both sense and antisense sequences). In both prokaryotes and eukaryotes, antisense RNA sequences are produced, but the functions of these RNAs are not entirely clear.[38] One proposal is that antisense RNAs are involved in regulating gene expression through RNA-RNA base pairing.[39]
A few DNA sequences in prokaryotes and eukaryotes, and more in plasmids and viruses, blur the distinction between sense and antisense strands by having overlapping genes.[40] In these cases, some DNA sequences do double duty, encoding one protein when read along one strand, and a second protein when read in the opposite direction along the other strand. In bacteria, this overlap may be involved in the regulation of gene transcription,[41] while in viruses, overlapping genes increase the amount of information that can be encoded within the small viral genome.[42]
Supercoiling
DNA can be twisted like a rope in a process called DNA supercoiling. With DNA in its "relaxed" state, a strand usually circles the axis of the double helix once every 10.4 base pairs, but if the DNA is twisted the strands become more tightly or more loosely wound.[43] If the DNA is twisted in the direction of the helix, this is positive supercoiling, and the bases are held more tightly together. If they are twisted in the opposite direction, this is negative supercoiling, and the bases come apart more easily. In nature, most DNA has slight negative supercoiling that is introduced by enzymes called topoisomerases.[44] These enzymes are also needed to relieve the twisting stresses introduced into DNA strands during processes such as transcription and DNA replication.[45]
Alternative DNA structures
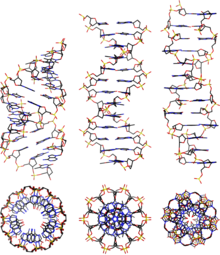
DNA exists in many possible conformations that include A-DNA, B-DNA, and Z-DNA forms, although only B-DNA and Z-DNA have been directly observed in functional organisms.[14] The conformation that DNA adopts depends on the hydration level, DNA sequence, the amount and direction of supercoiling, chemical modifications of the bases, the type and concentration of metal ions, and the presence of polyamines in solution.[46]
The first published reports of A-DNA X-ray diffraction patterns—and also B-DNA—used analyses based on Patterson functions that provided only a limited amount of structural information for oriented fibers of DNA.[47][48] An alternative analysis was proposed by Wilkins et al. in 1953 for the in vivo B-DNA X-ray diffraction-scattering patterns of highly hydrated DNA fibers in terms of squares of Bessel functions.[49] In the same journal, James Watson and Francis Crick presented their molecular modeling analysis of the DNA X-ray diffraction patterns to suggest that the structure was a double helix.[9]
Although the B-DNA form is most common under the conditions found in cells,[50] it is not a well-defined conformation but a family of related DNA conformations[51] that occur at the high hydration levels present in cells. Their corresponding X-ray diffraction and scattering patterns are characteristic of molecular paracrystals with a significant degree of disorder.[52][53]
Compared to B-DNA, the A-DNA form is a wider right-handed spiral, with a shallow, wide minor groove and a narrower, deeper major groove. The A form occurs under non-physiological conditions in partly dehydrated samples of DNA, while in the cell it may be produced in hybrid pairings of DNA and RNA strands, and in enzyme-DNA complexes.[54][55] Segments of DNA where the bases have been chemically modified by methylation may undergo a larger change in conformation and adopt the Z form. Here, the strands turn about the helical axis in a left-handed spiral, the opposite of the more common B form.[56] These unusual structures can be recognized by specific Z-DNA binding proteins and may be involved in the regulation of transcription.[57]
Alternative DNA chemistry
For many years, exobiologists have proposed the existence of a shadow biosphere, a postulated microbial biosphere of Earth that uses radically different biochemical and molecular processes than currently known life. One of the proposals was the existence of lifeforms that use arsenic instead of phosphorus in DNA. A report in 2010 of the possibility in the bacterium GFAJ-1 was announced,[58][59] though the research was disputed,[59][60] and evidence suggests the bacterium actively prevents the incorporation of arsenic into the DNA backbone and other biomolecules.[61]
Quadruplex structures
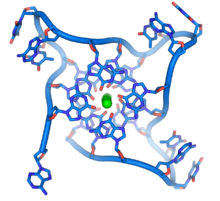
At the ends of the linear chromosomes are specialized regions of DNA called telomeres. The main function of these regions is to allow the cell to replicate chromosome ends using the enzyme telomerase, as the enzymes that normally replicate DNA cannot copy the extreme 3′ ends of chromosomes.[63] These specialized chromosome caps also help protect the DNA ends, and stop the DNA repair systems in the cell from treating them as damage to be corrected.[64] In human cells, telomeres are usually lengths of single-stranded DNA containing several thousand repeats of a simple TTAGGG sequence.[65]
These guanine-rich sequences may stabilize chromosome ends by forming structures of stacked sets of four-base units, rather than the usual base pairs found in other DNA molecules. Here, four guanine bases, known as a guanine tetrad, form a flat plate. These flat four-base units then stack on top of each other to form a stable G-quadruplex structure.[66] These structures are stabilized by hydrogen bonding between the edges of the bases and chelation of a metal ion in the centre of each four-base unit.[67] Other structures can also be formed, with the central set of four bases coming from either a single strand folded around the bases, or several different parallel strands, each contributing one base to the central structure.
In addition to these stacked structures, telomeres also form large loop structures called telomere loops, or T-loops. Here, the single-stranded DNA curls around in a long circle stabilized by telomere-binding proteins.[68] At the very end of the T-loop, the single-stranded telomere DNA is held onto a region of double-stranded DNA by the telomere strand disrupting the double-helical DNA and base pairing to one of the two strands. This triple-stranded structure is called a displacement loop or D-loop.[66]
|

|
| Single branch | Multiple branches |
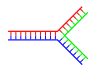
Branched DNA
In DNA, fraying occurs when non-complementary regions exist at the end of an otherwise complementary double-strand of DNA. However, branched DNA can occur if a third strand of DNA is introduced and contains adjoining regions able to hybridize with the frayed regions of the pre-existing double-strand. Although the simplest example of branched DNA involves only three strands of DNA, complexes involving additional strands and multiple branches are also possible.[69] Branched DNA can be used in nanotechnology to construct geometric shapes, see the section on uses in technology below.
Artificial bases
Several artificial nucleobases have been synthesized, and successfully incorporated in the eight-base DNA analogue named Hachimoji DNA. Dubbed S, B, P, and Z, these artificial bases are capable of bonding with each other in a predictable way (S–B and P–Z), maintain the double helix structure of DNA, and be transcribed to RNA. Their existence could be seen as an indication that there is nothing special about the four natural nucleobases that evolved on Earth.[70][71] On the other hand, DNA is tightly related to RNA which does not only act as a transcript of DNA but also performs as molecular machines many tasks in cells. For this purpose it has to fold into a structure. It has been shown that to allow to create all possible structures at least four bases are required for the corresponding RNA,[72] while a higher number is also possible but this would be against the natural principle of least effort.
Acidity
The phosphate groups of DNA give it similar acidic properties to phosphoric acid and it can be considered as a strong acid. It will be fully ionized at a normal cellular pH, releasing protons which leave behind negative charges on the phosphate groups. These negative charges protect DNA from breakdown by hydrolysis by repelling nucleophiles which could hydrolyze it.[73]
Macroscopic appearance

Pure DNA extracted from cells forms white, stringy clumps.[74]
Chemical modifications and altered DNA packaging
|

|

|
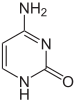
| cytosine | 5-methylcytosine | thymine |
Base modifications and DNA packaging
The expression of genes is influenced by how the DNA is packaged in chromosomes, in a structure called chromatin. Base modifications can be involved in packaging, with regions that have low or no gene expression usually containing high levels of methylation of cytosine bases. DNA packaging and its influence on gene expression can also occur by covalent modifications of the histone protein core around which DNA is wrapped in the chromatin structure or else by remodeling carried out by chromatin remodeling complexes (see Chromatin remodeling). There is, further, crosstalk between DNA methylation and histone modification, so they can coordinately affect chromatin and gene expression.[75]
For one example, cytosine methylation produces 5-methylcytosine, which is important for X-inactivation of chromosomes.[76] The average level of methylation varies between organisms—the worm Caenorhabditis elegans lacks cytosine methylation, while vertebrates have higher levels, with up to 1% of their DNA containing 5-methylcytosine.[77] Despite the importance of 5-methylcytosine, it can deaminate to leave a thymine base, so methylated cytosines are particularly prone to mutations.[78] Other base modifications include adenine methylation in bacteria, the presence of 5-hydroxymethylcytosine in the brain,[79] and the glycosylation of uracil to produce the "J-base" in kinetoplastids.[80][81]
Damage
DNA can be damaged by many sorts of mutagens, which change the DNA sequence. Mutagens include oxidizing agents, alkylating agents and also high-energy electromagnetic radiation such as ultraviolet light and X-rays. The type of DNA damage produced depends on the type of mutagen. For example, UV light can damage DNA by producing thymine dimers, which are cross-links between pyrimidine bases.[83] On the other hand, oxidants such as free radicals or hydrogen peroxide produce multiple forms of damage, including base modifications, particularly of guanosine, and double-strand breaks.[84] A typical human cell contains about 150,000 bases that have suffered oxidative damage.[85] Of these oxidative lesions, the most dangerous are double-strand breaks, as these are difficult to repair and can produce point mutations, insertions, deletions from the DNA sequence, and chromosomal translocations.[86] These mutations can cause cancer. Because of inherent limits in the DNA repair mechanisms, if humans lived long enough, they would all eventually develop cancer.[87][88] DNA damages that are naturally occurring, due to normal cellular processes that produce reactive oxygen species, the hydrolytic activities of cellular water, etc., also occur frequently. Although most of these damages are repaired, in any cell some DNA damage may remain despite the action of repair processes. These remaining DNA damages accumulate with age in mammalian postmitotic tissues. This accumulation appears to be an important underlying cause of aging.[89][90][91]
Many mutagens fit into the space between two adjacent base pairs, this is called intercalation. Most intercalators are aromatic and planar molecules; examples include ethidium bromide, acridines, daunomycin, and doxorubicin. For an intercalator to fit between base pairs, the bases must separate, distorting the DNA strands by unwinding of the double helix. This inhibits both transcription and DNA replication, causing toxicity and mutations.[92] As a result, DNA intercalators may be carcinogens, and in the case of thalidomide, a teratogen.[93] Others such as benzopyrene diol epoxide and aflatoxin form DNA adducts that induce errors in replication.[94] Nevertheless, due to their ability to inhibit DNA transcription and replication, other similar toxins are also used in chemotherapy to inhibit rapidly growing cancer cells.[95]
Biological functions
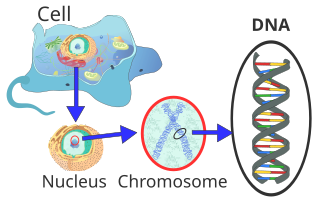
DNA usually occurs as linear chromosomes in eukaryotes, and circular chromosomes in prokaryotes. The set of chromosomes in a cell makes up its genome; the human genome has approximately 3 billion base pairs of DNA arranged into 46 chromosomes.[96] The information carried by DNA is held in the sequence of pieces of DNA called genes. Transmission of genetic information in genes is achieved via complementary base pairing. For example, in transcription, when a cell uses the information in a gene, the DNA sequence is copied into a complementary RNA sequence through the attraction between the DNA and the correct RNA nucleotides. Usually, this RNA copy is then used to make a matching protein sequence in a process called translation, which depends on the same interaction between RNA nucleotides. In an alternative fashion, a cell may copy its genetic information in a process called DNA replication. The details of these functions are covered in other articles; here the focus is on the interactions between DNA and other molecules that mediate the function of the genome.
Genes and genomes
Genomic DNA is tightly and orderly packed in the process called DNA condensation, to fit the small available volumes of the cell. In eukaryotes, DNA is located in the cell nucleus, with small amounts in mitochondria and chloroplasts. In prokaryotes, the DNA is held within an irregularly shaped body in the cytoplasm called the nucleoid.[97] The genetic information in a genome is held within genes, and the complete set of this information in an organism is called its genotype. A gene is a unit of heredity and is a region of DNA that influences a particular characteristic in an organism. Genes contain an open reading frame that can be transcribed, and regulatory sequences such as promoters and enhancers, which control transcription of the open reading frame.
In many species, only a small fraction of the total sequence of the genome encodes protein. For example, only about 1.5% of the human genome consists of protein-coding exons, with over 50% of human DNA consisting of non-coding repetitive sequences.[98] The reasons for the presence of so much noncoding DNA in eukaryotic genomes and the extraordinary differences in genome size, or C-value, among species, represent a long-standing puzzle known as the "C-value enigma".[99] However, some DNA sequences that do not code protein may still encode functional non-coding RNA molecules, which are involved in the regulation of gene expression.[100]

Some noncoding DNA sequences play structural roles in chromosomes. Telomeres and centromeres typically contain few genes but are important for the function and stability of chromosomes.[64][102] An abundant form of noncoding DNA in humans are pseudogenes, which are copies of genes that have been disabled by mutation.[103] These sequences are usually just molecular fossils, although they can occasionally serve as raw genetic material for the creation of new genes through the process of gene duplication and divergence.[104]
Transcription and translation
A gene is a sequence of DNA that contains genetic information and can influence the phenotype of an organism. Within a gene, the sequence of bases along a DNA strand defines a messenger RNA sequence, which then defines one or more protein sequences. The relationship between the nucleotide sequences of genes and the amino-acid sequences of proteins is determined by the rules of translation, known collectively as the genetic code. The genetic code consists of three-letter 'words' called codons formed from a sequence of three nucleotides (e.g. ACT, CAG, TTT).
In transcription, the codons of a gene are copied into messenger RNA by RNA polymerase. This RNA copy is then decoded by a ribosome that reads the RNA sequence by base-pairing the messenger RNA to transfer RNA, which carries amino acids. Since there are 4 bases in 3-letter combinations, there are 64 possible codons (43 combinations). These encode the twenty standard amino acids, giving most amino acids more than one possible codon. There are also three 'stop' or 'nonsense' codons signifying the end of the coding region; these are the TAG, TAA, and TGA codons, (UAG, UAA, and UGA on the mRNA).
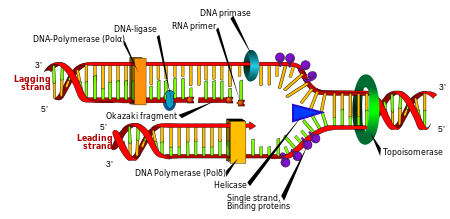
Replication
Cell division is essential for an organism to grow, but, when a cell divides, it must replicate the DNA in its genome so that the two daughter cells have the same genetic information as their parent. The double-stranded structure of DNA provides a simple mechanism for DNA replication. Here, the two strands are separated and then each strand's complementary DNA sequence is recreated by an enzyme called DNA polymerase. This enzyme makes the complementary strand by finding the correct base through complementary base pairing and bonding it onto the original strand. As DNA polymerases can only extend a DNA strand in a 5′ to 3′ direction, different mechanisms are used to copy the antiparallel strands of the double helix.[105] In this way, the base on the old strand dictates which base appears on the new strand, and the cell ends up with a perfect copy of its DNA.
Extracellular nucleic acids
Naked extracellular DNA (eDNA), most of it released by cell death, is nearly ubiquitous in the environment. Its concentration in soil may be as high as 2 μg/L, and its concentration in natural aquatic environments may be as high at 88 μg/L.[106] Various possible functions have been proposed for eDNA: it may be involved in horizontal gene transfer;[107] it may provide nutrients;[108] and it may act as a buffer to recruit or titrate ions or antibiotics.[109] Extracellular DNA acts as a functional extracellular matrix component in the biofilms of several bacterial species. It may act as a recognition factor to regulate the attachment and dispersal of specific cell types in the biofilm;[110] it may contribute to biofilm formation;[111] and it may contribute to the biofilm's physical strength and resistance to biological stress.[112]
Cell-free fetal DNA is found in the blood of the mother, and can be sequenced to determine a great deal of information about the developing fetus.[113]
Under the name of environmental DNA eDNA has seen increased use in the natural sciences as a survey tool for ecology, monitoring the movements and presence of species in water, air, or on land, and assessing an area's biodiversity.[114][115]
Neutrophil extracellular traps
Neutrophil extracellular traps (NETs) are networks of extracellular fibers, primarily composed of DNA, which allow neutrophils, a type of white blood cell, to kill extracellular pathogens while minimizing damage to the host cells.
Interactions with proteins
Zdroj:https://en.wikipedia.org?pojem=Single-stranded_DNAText je dostupný za podmienok Creative Commons Attribution/Share-Alike License 3.0 Unported; prípadne za ďalších podmienok. Podrobnejšie informácie nájdete na stránke Podmienky použitia.
Antropológia
Aplikované vedy
Bibliometria
Dejiny vedy
Encyklopédie
Filozofia vedy
Forenzné vedy
Humanitné vedy
Knižničná veda
Kryogenika
Kryptológia
Kulturológia
Literárna veda
Medzidisciplinárne oblasti
Metódy kvantitatívnej analýzy
Metavedy
Metodika
Text je dostupný za podmienok Creative
Commons Attribution/Share-Alike License 3.0 Unported; prípadne za ďalších
podmienok.
Podrobnejšie informácie nájdete na stránke Podmienky
použitia.
www.astronomia.sk | www.biologia.sk | www.botanika.sk | www.dejiny.sk | www.economy.sk | www.elektrotechnika.sk | www.estetika.sk | www.farmakologia.sk | www.filozofia.sk | Fyzika | www.futurologia.sk | www.genetika.sk | www.chemia.sk | www.lingvistika.sk | www.politologia.sk | www.psychologia.sk | www.sexuologia.sk | www.sociologia.sk | www.veda.sk I www.zoologia.sk

